NEWSFLASH To download our “Recommendations for using automatic bat identification software with full spectrum recordings” written with Paola Reason and Kate Jones Click HERE.
To download a visual guide to the sound identification of European bats click HERE.
Passive real-time detectors are triggered when they detect sound within a certain frequency range. Monitoring on this scale can generate a very large volume of recordings, efficient processing of which is greatly aided by a semi-automated approach for assigning recordings to species.
For our projects we make use of an acoustic classifier for UK bats developed in collaboration with the Muséum National d’Histoire Naturelle in Paris using TADARIDA (a Toolbox for Animal Detection in Acoustic Recordings Integrating Discriminant Analysis; Bas 2016; Bas et al. 2017).
STEP 1 – UK BAT CLASSIFIER
Recordings are initially passed through the TADARIDA random forest classifier (Step 1). This entails extraction of 150 measures of call characteristics from each recording (Bas et al. 2017), and a comparison of these against measurements taken from an extensive reference library of manually identified ultrasound recordings. The initial reference library of recordings was provided by Muséum National d’Histoire Naturelle, but using TADARIDA we have been able to continue to work on a UK classifier for bats (and other species groups – see publications), and to add to and share reference libraries.
The classifier allows up to four different “identities” to be assigned to a single recording, according to probability distributions between detected and classified sound events. From these, species identities are assigned by the classifier, along with an estimated probability of correct classification (as compared with the underlying training database) on a scale of 0–1. For common Pipistrellus pipistrellus and soprano pipistrelle Pipistrellus pygmaeus, which typically account for >95% of all bat recordings, the call shape (similar to a hockey-stick) and frequencies of common and soprano pipistrelle are sufficiently characteristic to allow reliable classification of these species by the classifier. For these species, TADARIDA identifications for which the estimated probability of correct classification was high (≥ 0.8), were taken as being accurate [but note that we are currently carrying out analysis to formally quantify accuracy for each species, and to define more objective thresholds].
STEP 2 – MANUAL CHECKING OF SPECTROGRAMS
Manual checking (Step 2) of spectrograms using software SonoBat (http://sonobat.com) was used as an independent check of the original species identities assigned by the TADARIDA classifier. Because the Tadarida classifier is able to detect and identify weaker bat calls than SonoBat is able to show, an additional viewer Audacity was sometimes used to check the identify of recordings, where there was no obvious confusion species. Using the output from Step 1, manual checks were carried out on a random sample of 1,000 recordings each of common and soprano pipistrelle, to verify that classifier identification of these species was accurate. For the other species, we inspected all recordings with SonoBat regardless of the associated probability of correct classification. Species identities were checked (and re-classified if necessary) based on call parameters defined in Russ (2012) and Barataud (2015).
STEP 3 – CONSIDERING SEQUENCES OF RECORDINGS
Once species identities had been checked by looking at individual recordings in isolation, calls assigned to species whose calls had the most potential to be confused with those of other species (e.g. bats in the genus Myotis and Nyctalus) were re-examined in SonoBat, comparing them to other recordings potentially of the same bat made from the same location on the same night at neighbouring points in time (Step 3). All subsequent analyses use final identities upon completion of the above inspection and (where necessary) correction steps.
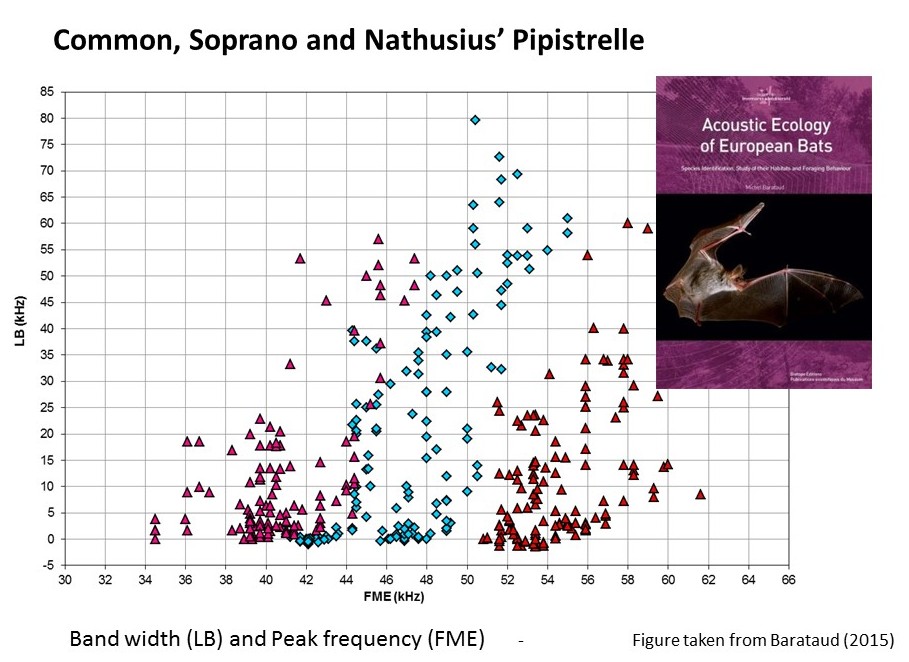
Figure 1 below provides an example of how combining peak frequency and bandwidth in an analyses for Common, Soprano and Nathusius’ Pipistrelle, would provide better discrimination than considering peak frequency alone. In practice our classifier (Step 1) considers the potential of a wider range of call parameters for improving discrimination. Recordings of Whiskered Myotis mystacinus or Brandt’s bat Myotis brandtii are currently treated as a species pair. Feeding into this process, we have spent time in Paris with colleagues at the Muséum National d’Histoire Naturelle, to learn from their experience in the acoustic identification of bats, and considering the scarcity of records of Whiskered and Brandt’s bats from Norfolk and Scotland prior to this study, to ensure that they would confidently assign the same recordings to this species pair. Currently about 72% of recordings initially assigned to Whiskered / Brandt’s in Step 1 are reassigned to genus (Myotis species), but we accept that identification of all Myotis is challenging, and we recommend that identification is followed up where possible with more targeted (e.g mist-netting) to ground truth identification. Information on the number of recordings removed or reassigned to genus or species at each step of this process for the first two years of the Norfolk survey can be viewed in Table 2 (from Newson et al. 2015). For more information on the steps involved as applied to the first two years of Norfolk data and first outputs to come out of the project see Newson et al. (2015).